The global pandemic of COVID-19 disease has affected the lives of millions of people around the world. Numerous research teams dealing with the health research have radically changed their plans in order to devote their capacity to research the disease with the purpose to find effective help as soon as possible.
Interesting initiative was started by researchers from the Loschmidt Laboratories of the Faculty of Science of Masaryk University (MUNI), the Institute of Computer Science of Computer Science at MUNI, the RECETOX MUNI Research Center and the International Center for Clinical Research at the University Hospital at St. Anny in Brno (FNUSA-ICRC).
Their contribution to understanding the virus envisages the use of computational biochemistry, artificial intelligence and the principles of machine learning.
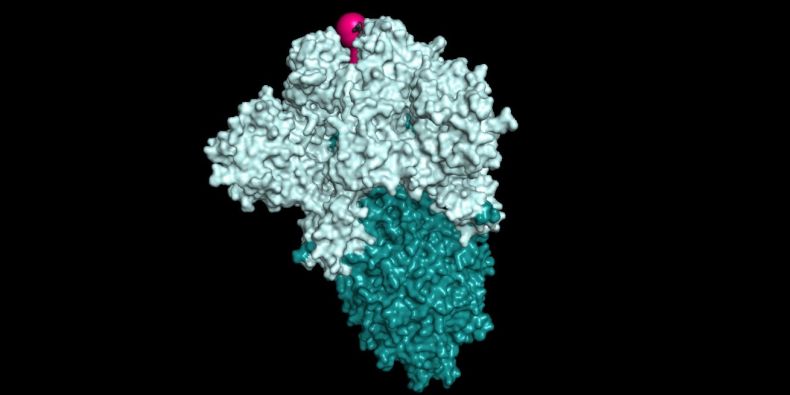
"Using our own CaverDock software, we focused on the computer study of a protein that is key in the spread of SARS-CoV-2 virus in the human body," described Jiří Damborský from the Faculty of Science MU and head of the FNUSA-ICRC Protein Engineering research team. It is a viral glycoprotein S whose trimer (a molecule of three monomers) forms the protrusions of the SARS-CoV-2 coronavirus envelope and binds to human host cells.
The researchers performed so-called virtual screening of 4,359 already approved drugs to find out their effectiveness on this particular protein.
"For the first time, we used the CaverDock program we are developing to study such a large number of molecules. The program proved excellent, practically 100% robustness, and thus became one of the most reliable tools in its category, " said the author of the algorithms Jiří Filipovič from the Institute of Computer Science at MUNI. The CaverDock program was developed thanks to the support of the MUNI Internal grant agency financing interdisciplinary research and is provided to a wide user community by the national infrastructure ELIXIR.CZ.
"We performed several simulations of changes in the molecular structure of this protein to find out which of the known drugs could have the greatest efficacy," said Gaspar Pinto of Loschmidt Laboratories MUNI and FNUSA-ICRC. Because a similar process generates such an enormous amount of data, machine learning methods and artificial intelligence are used to analyze them. "We also submitted a grant application to Microsoft's Azure cloud program," Pinto added.
Based on these calculations, several approved drugs that can block the function of this protein and thus prevent the virus from binding to the human host cell have been identified. "Artificial intelligence can also offer new drug structures that bind to protein even more effectively," Pinto said. According to his opinion, this is a new area of COVID-19 disease research, where software solutions are being established to accelerate the development of new drugs.